Research Projects
Spore Germination and Infection by Pathogenic and Endophytic Fungi
Spore germination stages of Fusarium graminearum and Metarhizium anisopliae.
The spore germination is the initial stage of fungal colonization on plants. The information on genes that govern the initial growth stages is vital for developing efficient strategies to prevent fungal diseases on plants. Our research focuses on understanding the conserved and unique gene expression pattern of spore germination between a plant pathogenic fungus, Fusarium graminearum and an endophytic fungus, Metarhizium anisopliae. We are working on identifying the genes implicated in spore germination by performing transcriptome analysis and functional validation assays using targeted gene deletion mutants.
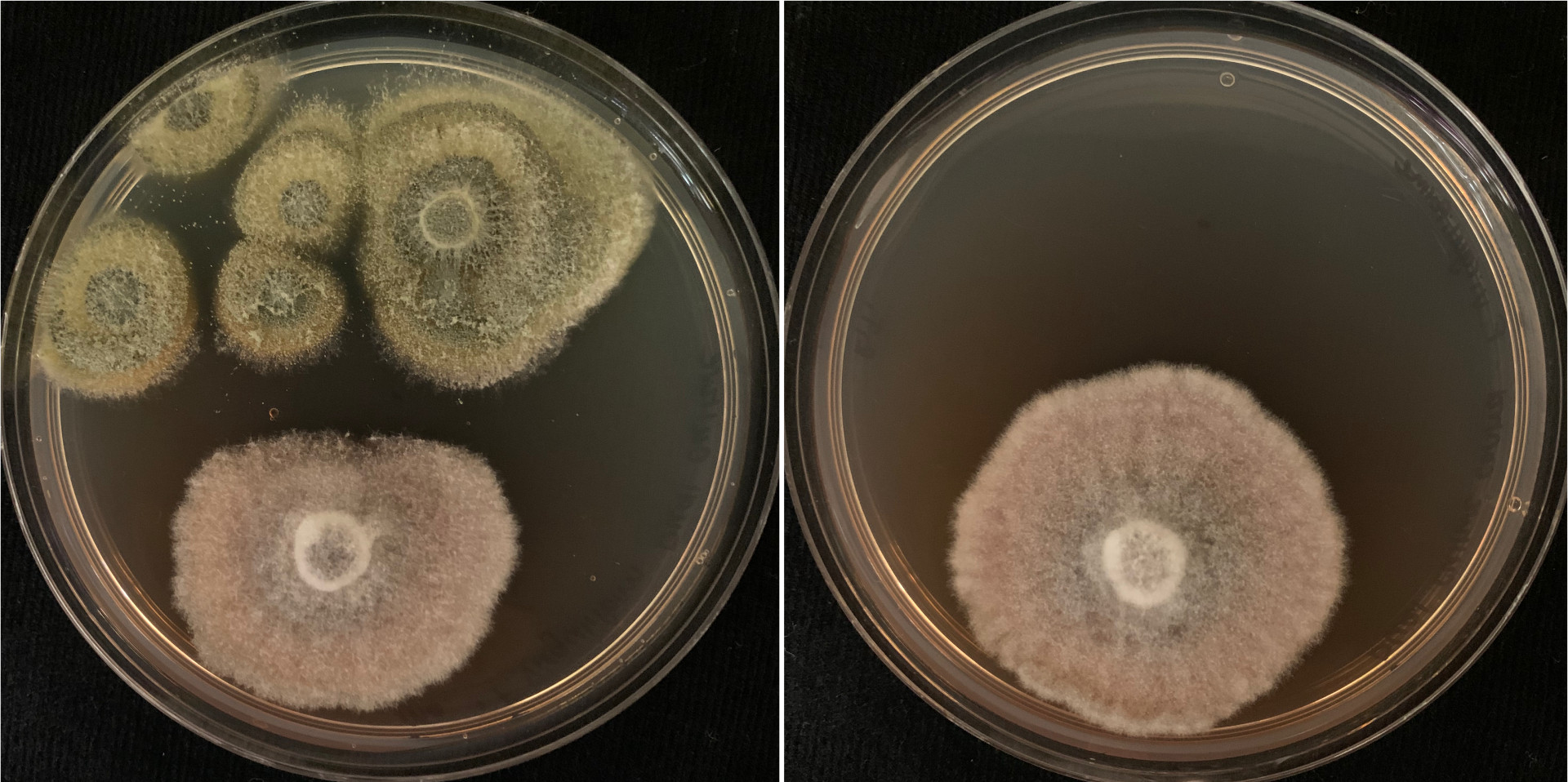
Spore Development and Forcible Discharge in Fusarium graminearum
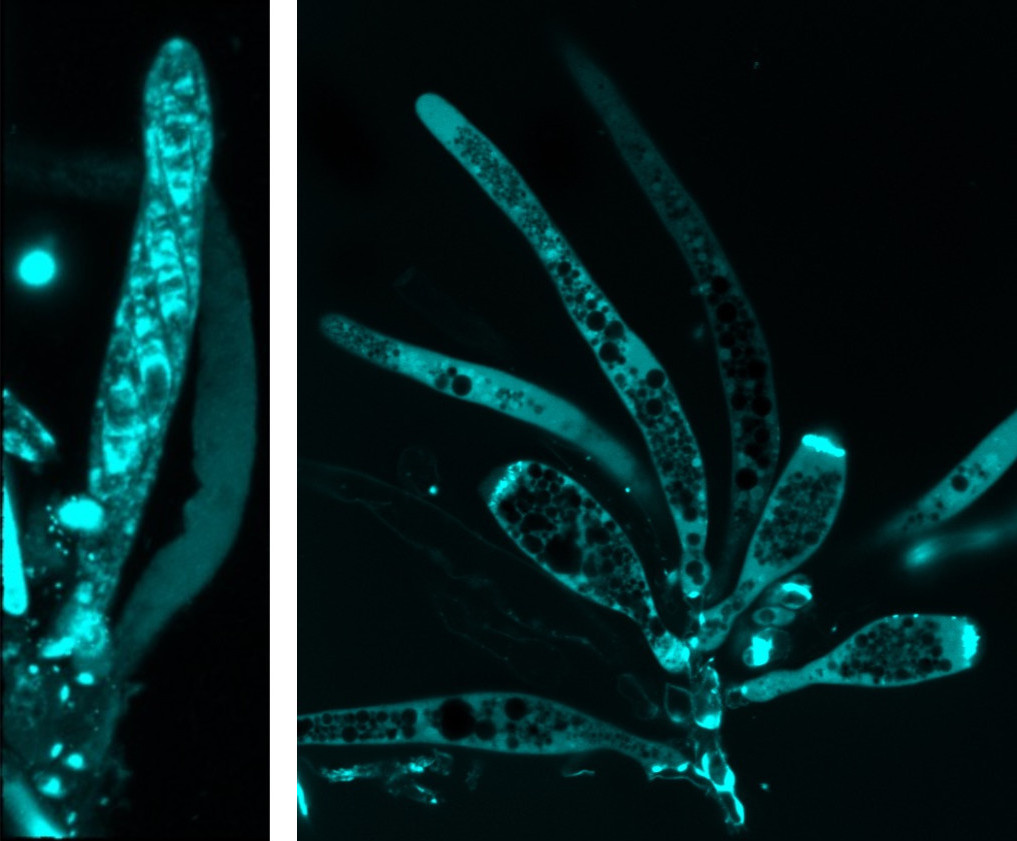
Left: Mature ascus of wild type. The cell walls of the ascus and the maturing spores are stained with Calcofluor white. The spores in the empty ascus visible in the background to the right have already been discharged. Right: Mature (elongated) and immature (rotund) asci of a developmental mutant. The cell walls are stained with Calcofluor white for visibility. The lack of spores in the mature asci indicate defects in ascus development and spore formation in the mutant.
Forcibly shooting spores from a pressurized ascus is a major contributor to the initiation and spread of many of the most important and devastating agricultural diseases worldwide. Since 1996, the Trail Lab has been seeking to understand the processes necessary of the development of fruiting bodies and the mechanisms employed in forcible dispersal of ascospores, which play an essential role in the seasonal initiation and spread of Fusarium Head Blight disease. We have applied functional genetics to identify genes essential for spore firing. Currently we are taking a closer look at the minute details of ascus structure. We are comparing ascus structure in mutants that no long fire their spores to the wild-type asci.
Our efforts to tease apart the various processes contributing to forcible spore discharge include the study of physiological processes such as the buildup of osmoticum and influx of water, the use of reverse genetics where we have identified several candidate genes such as Cch1 and Mid1 that play critical roles in spore discharge and the use of several mutants generated in our lab that show a discharge minus phenotype. One of these mutants produces rotund asci that elongate as they mature. It appears to be defective in cell wall formation and spore development.
Evolution of the Transcriptome Regulating Spore Development and Germination in Opportunistic Human Pathogens

Left: Spores emerging from phialides, growing out of hyphae in Fusarium oxysporum. Scanning Electron Microscopy reveals the formation of microconidia from these specialized cells. Left, early spore emergence from phialide; Right: mature spore falling out of the phialide.
We are investigating transcriptional dynamics of orthologs across five phylogenetically diverse Ascomycota species during four contiguous stages of spore germination. This has led to the identification of genes with significantly evolved expression in opportunistic human pathogens versus nonpathogenic relatives. We identified a transcription regulator, abaA, that plays a crucial role in conidiation among the species, and through targeted experimentation, we have revealed that the changes in gene expression of abaA has impacts on the development of conidia by phialides of each species.